Sherman Silber
Infertility Center of St. Louis, St. Luke’s Hospital, St. Louis, Missouri
Fertility and Sterility, June 30, 2011
Download PDF version of this article
The Y chromosome contains 60 multicopy genes composed of nine different gene families concentrated in regions of multiple repeat sequences called amplicons arranged in mirror images called palindromes. This pattern is susceptible to deletions caused by homologous recombination with itself, and can explain the presence of small numbers of sperm in otherwise azoospermic men.
The Y chromosome contains 60 multicopy genes composed of nine different gene families concentrated in regions of multiple repeat sequences called amplicons arranged in mirror images called palindromes. This pattern is susceptible to deletions caused by homologous recombination with itself, and can explain the presence of small numbers of sperm in otherwise azoospermic men.
Almost concurrently with the development of intracytoplasmic sperm injection (ICSI) and testicular sperm extraction (TESE) for azoospermia in 1993, we began to study the genetic causes of male infertility via the first mapping of the Y chromosome in azoospermic males and in fertile control male populations, eventually leading to complete sequencing of the Y chromosme. This led to an understanding of why tiny amounts of sperm are often found in the testis of azoospermic men previously thought to be making no sperm (1–8). Subsequently, most of the research into the genetics of male infertility has been focused on aberrations of the Y chromosome. This specialized male chromosome, the Y chromosome, contains many genes that are involved in spermatogenesis, concentrated in a peculiar pattern of nucleotide repeats and mirror image inversions called amplicons and palindromes. Deletions involving these regions of the Y are found in 15% of severely infertile males and have been shown to be transmitted to male offspring via ICSI, presumably causing fertility problems in these children later in life (5, 9, 10).
The sequence of the Y also gives us a perspective about other male infertility genes that are widespread throughout the genome and which also could transmit infertility to future generations of ICSI offspring. More importantly perhaps, understanding the Y chromosome helps us to comprehend why men who are seemingly azoospermic often have some residual tiny amount of spermatogenesis that can be used for successful ICSI (11–13). Many genes on the Y chromosome required for spermatogenesis have multiple copies and even ”backup” homologues elsewhere in the genome, such asDAZL and CDYL (14, 15). Another broader scientific benefit is that it has amplified our understanding of sex determination in general and has even helped to explain the origin of Turner syndrome (16). Indeed, studies of male infertility and the Y chromosome have gone as far as showing why XO female patients with Turner syndrome are generally caused not by the loss of an X chromosome in the female embryo, but rather by a loss of the Y chromosome in a male embryo.
TREATMENT OF MALE INFERTILITY
Until 2 decades ago, there were no good treatment options for infertile couples when the male had severely impaired spermatogenesis. In fact, there are still no clinical therapies to correct deficient spermatogenesis (17–30). Since the introduction of TESE and ICSI by us and the Brussels Dutch-Speaking Free University in 1992, however, there has been a revolution in our thinking about male infertility (31, 32). Infertile couples with the most severe cases of male infertility, even with apparently 100% abnormal morphology and/or only rare spermatozoa in the ejaculate, can now have pregnancy and delivery rates not apparently different from conventional IVF with normal sperm (33–35).
In 1993, we first introduced microsurgical epididymal sperm aspiration (MESA) in conjunction with ICSI for the treatment of obstructive azoospermia (3, 12, 36–38). A few months later, TESE was also found to be effective for the majority of cases of nonobstructive azoospermia as well (3, 12, 13, 38, 39). The reason is that ~60% of azoospermic men with presumably no sperm production actually do have a minute amount of sperm production in the testis that is not quantitatively sufficient to spill over into the ejaculate, but which is adequate for ICSI (11, 13, 27, 39–46). Thus, even men with extremely low sperm counts, or with spermatogenesis so deficient in quantity that no sperm at all can reach the ejaculate, could now have children with the use of TESE-ICSI (3, 10, 47).
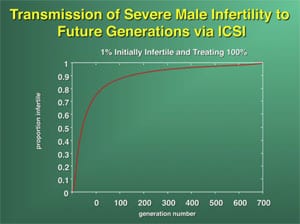
It is with these cases of nonobstructive azoospermia and severe oligozoospermia that the greatest concern has been registered for the fertility of future generations. If severe oligozoospermia or azoospermia is of genetic origin, then ICSI may create a potential problem of proliferation of male infertility in future generations (48). In fact, if one-half of all azoospermic men were to undergo ICSI, the incidence of male infertility would double within seven generations (Supplemental Fig. 1) (49).
GENETIC CAUSES OF MALE INFERTILITY: ROLE OF THE Y CHROMOSOME
Karyotyping reveals that structural chromosomal abnormalities, such as translocations and inversions, are found in about 1% of azoospermic men, but the most common karyotypic chromosomal abnormalities in azoospermic men (2%) involve the sex chromosomes, such as in Klinefelter syndrome (47,XXY) (50–64). In the majority of these cases, surprisingly, we can usually find a few rare sperm adequate for fertilization using ICSI (44, 65–67). (See the Supplemental Material for more)
Suspicion of involvement of the Y chromosome in male infertility actually arose originally from cytogenetic evidence reported more than 35 years ago (68). This karyotyping study showed grossly obvious terminal Y chromosome deletions in a very small percentage (0.5%) of azoospermic men who were otherwise phenotypically normal. It was then postulated that the Y chromosome contained a socalled Azoospermia Factor gene (AZF). During the middle 1990s, we showed that the long arm of the Y chromosome contains not one but many distinct deletion intervals and at least 60 genes belonging to nine gene families whose exclusive functions are in spermatogenesis (1–4, 6–8, 69). Interestingly, several of these multicopy spermatogenesis genes on the Y chromosome have ”backup” homologues elsewhere in the genome, like DAZLon chromosome 3 and CDYL on chromosome 6.
In fact, the deletion frequency of one or more of these regions on the Y chromosome in men with azoospermia or severe oligozoospermia is ~15% (Fig. 1) (1, 3, 70). After our initial report, many laboratories throughout the world have reported on these submicroscopic deletions of the Y chromosome in azoospermic and severely oligozoospermic men (25, 69, 71–101). In fact, deletion screening of the Y chromosome is now considered standard practice for severely oligozoospermic and azoospermic patients undergoing assisted reproduction in most countries in the world.
AZFa
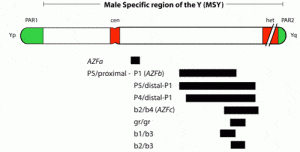
The AZFa region differs from the AZFb and AZFc regions because of its nonrepetitive structure and its low deletion frequency (Fig. 2). AZFa deletions are very uncommon and only a few rare patients have been described (69, 79, 87, 102–104). However, studies of this region are highly useful in understanding the genetic basis of male infertility. The AZFaregion spans ~800 kilobases (kb) and contains two functional single-copy genes: USP9Y and DBY (104, 105). This mutation represented the first and only case of a point mutation rather than a large microdeletion causing a single gene defect on the Y chromosome associated with spermatogenic failure (104). The lack of sequence repeats which characterize the rest of the Y chromosome made this particular region of the Y amenable to such a point mutation search. Because almost all other spermatogenesis genes on the Y chromosome, such as those in AZFb and AZFc, are multicopy, searching for point mutations in these genes is virtually impossible. It is also questionable whether point mutations in a single gene of a gene family will give rise to a severe infertility phenotype, because the remaining intact copies of the gene could potentially compensate for the loss of function of the mutated gene. This is an important lesson for why spermatogenic defects come in varying degrees of severity and why even in azoospermic men there are usually still a tiny number of healthy sperm ”hiding” in the testes.
The AZFa region provides a good model for the interaction and overlapping functions of multiple genes and sheds light on the polygenic nature of the genetic control of spermatogenesis. When the entire AZFa region is deleted, taking out bothDBY and USP9Y, the spermatogenic defect is severe and the patient is always azoospermic. In contrast, when only one gene is affected, such as loss of USP9Y function owing to a specific point mutation, this results in a less severe phenotype of maturation arrest with a few pachytene spermatocytes developing into a few mature sperm in some seminiferous tubules. Thus, the loss of DBY (the only other gene in the AZFa region) likely exacerbates the spermatogenic consequences of the loss of USP9Y. (See the Supplemental Material for more).
AZFb
Deletions of the AZFb region are slightly more common than deletions of the AZFa region but are still found in a very small percentage of azoospermic men (69, 95, 106–108). Interestingly, all men with deletions of AZFb described to date are azoospermic and show complete absence of spermatozoa in the testis (14, 69, 71, 81, 106, 107–111). Therefore, similarly to AZFa deletions, there are no reports on transmission of an AZFb deletion to ICSI offspring. Many functionally active genes are clustered within the AZFb region (81, 112, 113). Because of the presence of multiple sequence repeats within this region, in contrast to AZFa, efforts to define the AZFb region precisely had been hampered, but now the exact content and extent of the AZFb region has been elucidated and the entire Y chromosome has been fully sequenced (8, 106).
In fact, the region frequently referred to simplistically as AZFb overlaps AZFc and has a very large number of repeat copies of genes and pseudogenes (RBMY, PRY, TTTY) arranged in a complex of palindromes (inverted repeat sequences). That is why we technically object to this commonly used terminology of ”AZFb.” We prefer the more specific palindromic terminology. Deletions in this region of the Y chromosome are extensive and are much less common than in AZFc, and they are caused by weak ”breakpoints” in the center of its inverted repeats. This is a less common deletion mechanism than that which plagues the AZFc region.
AZFc
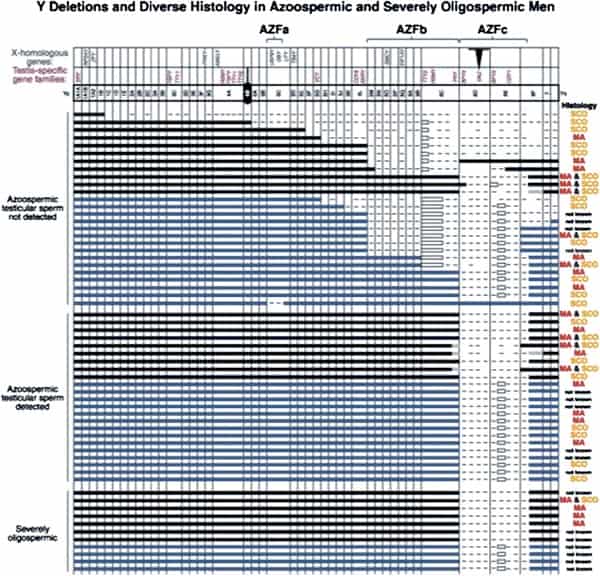
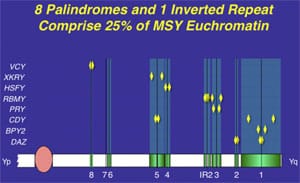
The most commonly deleted and best-studied region on the Y chromosome is the AZFc region. Deletion of the AZFcregion is found in ~12% of azoospermic men and in 6% of severely oligozoospermic men (1–3, 70). The complete nucleotide sequence of the AZFc region reveals an extraordinary structure and genetic composition. The region is constructed from very large areas of absolute sequence identity called amplicons which are arranged in direct repeats, inverted repeats, or palindromes. The AZFc region spans 3.5 Mb (huge but not quite as huge as AZFb with its length of 8 Mb) and contains seven separate families of genes with a total of 19 genes that are all exclusively expressed in the testis (Fig. 3). This so-called ”microdeletion,” AZFc, is actually a huge section of DNA, but not huge enough to show up with karyotyping. Interestingly, absence of this large 3.5-Mb AZFc chunk of the Y chromosome seems to have no other deleterious effect except upon spermatogenesis, exemplifying the remarkably specialized function of this region of the Y chromosome (2). These genes only affect spermatogenesis and nothing else.
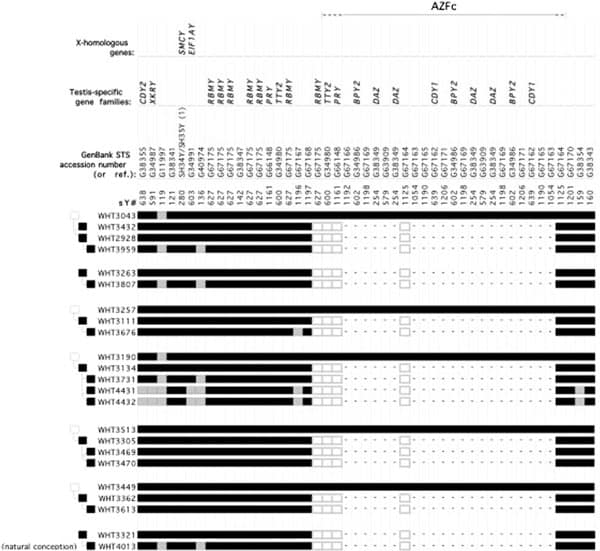
The DAZ gene family, which is one of the seven gene families located within AZFc, was one of the first spermatogenesis genes identified on the human Y chromosome and comes in four almost identical copies (1, 6). The human DAZ genes were shown to be transcribed specifically in spermatogonia and in early primary spermatocytes (114). Interestingly, homologues of DAZ in other species were also shown to be involved in control of spermatogenesis, supporting an essential role of this gene in male fertility in humans as well as almost all other animals. Homologues of DAZ have been found inDrosophila (termed boule), in mice (termed Dazl), in frogs (termed Xdazl), and even in worms (termed daz-1) (115–120). Therefore, DAZ is the most ancient and well conserved spermatogenesis gene. In contrast to its presence in humans, theDAZ gene in these other species is single copy and located on an autosome rather than on the Y chromosome. In the human, DAZ is present on the Y chromosome in four near-identical repeat copies (99.9% homology) arranged in two clusters with two genes in each cluster (2, 7).
The human also retains an autosomal homologue of DAZ called DAZL, which is located on chromosome 3 (6). During evolution, some time after the split of Old and New World monkeys ~30 million years ago, the DAZLgene was transposed to the Y chromosome like a simple translocation event (Supplemental Fig. 2). Once autosomal DAZL was transposed from chromosome 3 to Y, it was amplified and pruned until it became the modern-day DAZ gene family of four copies on the Y and still two autosomal copies on chromosome 3. In fact there is yet another DAZfamily homologue in humans, termed BOULE, that resembles the fly homologue boule, even more closely than DAZ or DAZL (121). The exact interaction and possible functional overlap between these three members of this interesting gene family holds the clue to the peculiar finding of a few surviving sperm in the testes of azoospermic men because of genetic ”backup.” In fact, >80% of azoospermic men with an AZFc deletion have some sperm in the testis retrievable with TESE (4, 9, 10).
The DAZL gene on chromosome 3 was the original human DAZ gene, which jumped over to the Y chromosome 30 million years ago and resulted in our modern Y chromosome DAZ gene. The Y chromosome, with its lack of recombination during meiosis, represents a safe harbor for the proliferation of genes that are beneficial to the male but detrimental to the female. This is how the Y chromosome, over evolutionary time, tends to accumulate genes from throughout the entire rest of the genome that control spermatogenesis.

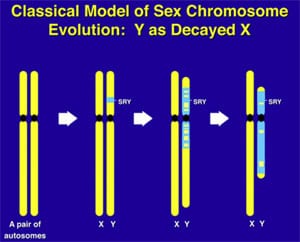
The Y chromosome in mammals, and other sex-determining chromosomes such as the W chromosome in birds, evolved differently over and over again in different genera, always with totally different genes and nucleotide sequences, but always with the same general theme: a degradation of its X homologues owing to failure of recombination (Supplemental Fig. 3, available online at www.fertstert.org) and at the same time an accumulation and amplification of multiple copies of male-specific spermatogenesis genes from all over the genome. It is this multiplicity of gene copies all of which contribute quantitatively to total sperm production that explains the minute amount of sperm we can find at TESE in azoospermic men, because of ”res- cue” from the continued presence of at least one of these gene copies (such as DAZLon chromosome 3) even though most are missing or deleted.
Deletions of the entire AZFc region result in loss of all four DAZ copies. Recent reports however, indicate that deletions involving only some of the DAZ genes are also found in infertile men, but these smaller deletions are found in men with only mild oligozoospermia, indicating a possible gene dosage effect, i.e., men with a deletion of only two DAZ genes are less affected than men with a deletion of all four copies (122–126). These are called GR-GR deletions, that is, half of AZFcis deleted. There are also less studied AZFc deletions, such as B1-B3 and B2-B3. These data illustrate that infertility is a complex multigenic disorder and that disruption of different genes or disruption of some genes of a gene family can result in different degrees of spermatogenic failure.
MECHANISM OF DE NOVO Y CHROMOSOME DELETIONS
Research into deletions involving AZFa and AZFc has provided interesting data on the mechanism of deletions, which sheds light on the inexorable decline in spermatogenesis in humans and in other species which have no ”sperm competition.” Deletions on the Y chromosome are caused by ”illegitimate” homologous recombination between highly similar or identical sequences which are found on the Y in great abundance. For example, homologous recombination between two identical sequence stretches results in dropout of the intervening AZFa region (105, 127, 128). Because these sequences of identical nucleotide repeats are shorter than those in AZFc, AZFa deletions are very rare.
However, for AZFc the substrates for homologous recombination are two repeats that are >99.9% identical and as long as 229 kb in length (2). The frequency with which these deletions occur seems to correspond to the length of the stretch of homology. Therefore, deletions of AZFc, caused by homologous recombination between 229-kb repeats, are far more common than deletions of AZFa, which are caused by repeats of only 10 kb in length. The very repetitive nature of the Y chromosome seems to be the cause of its instability over an evolutionary time frame, as well as in our current infertile male patient population, but it is also the method that the Y has adopted (albeit inefficient) for its survival, called ”gene conversion” (129).
What is equally fascinating is how we survived at all as our X homologous genes on the Y chromosome deteriorated because of failure of meiosis with its nonrecombining mate (130, 131). In fact, this deterioration of X-homologous Y genes was the whole reason for the evolution of ”X inactivation,” to put males and females on an equal footing despite different gene dosages.
EVOLUTION AND GENETIC CONSTITUTION OF THE HUMAN Y CHROMOSOME
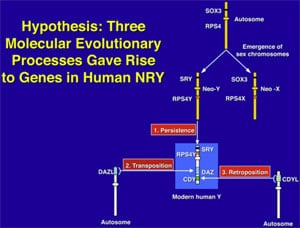
What makes the Y chromosome, with its confounding repeats, polymorphisms, and degenerating regions such an interesting object of study for male infertility? The answer lies in the evolutionary history of the X and Y chromosomes. Over the course of the past 240–320 million years of mammalian evolution, the X and Y chromosomes have evolved from what was originally a pair of ordinary autosomes (Supplemental Fig. 3) (14, 130–137). During that evolution, just as most of the ancestral X genes were decaying on the Y because of the lack of meiotic recombination, genes that control spermatogenesis arrived on the Y from autosomes. Once on the Y, these formerly autosomal genes amplified into multiple copies and achieved greater prominence through the process called ”gene conversion” (6, 129, 138). Spermatogenesis genes that arrived on the Y, but came originally from autosomes, include the DAZ (from autosome 3) and CDY (from autosome 6) genes which are among the seven gene families located in AZFc (Supplemental Fig. 4) (6, 15). Other spermatogenesis genes on the Y, such as RBMY, have persisted in their original position as on the X (139–142). The ancestral gene that remained on the X chromosome (RBMX) retained its widespread cellular functions, whereas RBMY, which persisted on the receding Y chromosome, evolved a male-specific function in spermatogenesis (141, 143–145). Male-benefit genes have thus arrived and accumulated on the evolving Y chromosome over many millions of years via three mechanisms: 1) persistence of genes on the ancestral X that evolved a male-specific function (RBM to RBMY); 2) retroposition from an autosome via reverse transcription (CDL to CDY); and 3) transposition from an autosome via translocation (DAZL to DAZ; Supplemental Fig. 2).
This evolution of the modern X and Y chromosomes was initiated by the emergence of a male sex-determining gene (now known as SRY) on what was originally an ordinary pair of autosomes (Supplemental Fig. 3) (15, 136, 137, 141). Genes associated with the nonrecombining SRY region that were specifically beneficial for male function or antagonistic to female function, flourished on the evolving Y chromosome despite the deterioration of more generalized genes that lacked the DNA repair benefits of meiosis (145–150).
The next question that the evolution of the X and Y chromosomes presents, which is crucial for understanding male infertility and ICSI, is how this degraded Y chromosome survives at all. The lack of recombination of the male-determining Y chromosome has led to complete deterioration and loss of most of its original 1,438 genes (the number of genes on its corresponding X) and an accumulation of only 60 genes (only nine gene families) which are male specific and are located in areas of sequence identity that promotes further deletions. So how does the Y survive at all, and why do we humans retain any spermatogenesis at all?
The answer is ”gene conversion” (129). It answers the question of how these ampliconic repeats and palindromic inversions occur. When autosomes recombine during meiosis, DNA is exchanged in a way that the accumulated mutational errors of life get corrected in the germ cells via this DNA exchange. In a sense, the autosomes ”have sex” with each other. This correctional meiosis cannot occur with the Y chromosome. Instead, the Y chromosome ”has sex” with itself. That is, the like-sequence repeats of the Y chromosome ”recombine,” in a sense, with each other, an ”illegitimate” homologous recombination. This ”gene conversion” creates and repairs the multiple copies and inverted DNA sequence repeats that characterize the Y and indeed all sex-determining chromosomes. So if there is a miss- ing or deleted copy of a spermatogenesis gene, there are other backup copies that can still rescue spermatogenesis to some degree.
Throughout our whole study of Y deletions, the most confusing was the so-called ”terminal deletion,” shown in the top six cases of Figure 1. With molecular analysis, they appeared with initial mapping not to be interstitial deletions, but rather ”isodicentric,” meaning that the Y chromosome did not just come to an end point proximally, but rather was doubled over on itself with like copies annealed to each other. This is caused by a kind of ”gene conversion” that went awry. (See the Supplemental Text for more.) Thus, ”gene conversion” as a mechanism for maintenance and repair of the Y chromosome has more limited value than the standard meiosis that the X chromosome in the ovary and the autosomes enjoy.
THE Y CHROMOSOME AND SPERMATOGENESIS IN HUMANS AND IN APES
Comparing spermatogenesis in humans, chimpanzees, and gorillas has always been fascinating (52, 149). Chimpanzees, which weigh only about 100 pounds, have enormous 8-cm-diameter round (not oval) testes with sperm counts of >1 billion/mL. Yet gorillas, which weigh as much as 600 pounds or more, have tiny testes, very poor spermatogenesis, and, in the sparse literature on gorilla testicular histology, in the majority of cases have what appears to be Sertoli cells only (151). Humans, the closest living relatives to chimpanzees and gorillas, fall somewhere in between.
Most intriguing is to compare the human Y chromosome to the chimpanzee Y chromosome, both of which have been fully and accurately sequenced (8, 152). Unfortunately, the gorilla Y has not yet been sequenced. Some interesting differences are noted between the human and chimpanzee Y chromosomes (152). The chimpanzee Y chromosome has many more amplicons and palindromes than the human, but it nonetheless has much fewer ampliconic genes (25 compared with 60). So the increased sperm production of chimpanzees cannot be explained by the presence of more testis-specific genes on their Y chromosome. But interestingly, the chimpanzee Y is missing a gene (PRY) that is present on AZFc in two copies in the human. You cannot attribute the vast superiority of chimpanzee spermatogenesis to having more copies of spermatogenesis genes. But the PRY gene, which humans have and chimpanzees do not have, could be a suppressor of spermatogenesis. Thus, a deletion of PRY in the chimpanzee could conceivably be a reason for its huge amount of spermatogenesis. Furthermore, it is fascinating that this gene has also been found to be present in only one copy in rare humans who have incredibly high sperm counts, approaching half a billion (153). Thus, comparing the superfertile chimpanzee Y chromosome to its less fertile human cousin, can help us to better understand the genetic control of spermatogenesis in our infertile male patients.
OTHER MALE INFERTILITY GENES NOT ON THE Y CHROMOSOME
Although most research into the genetics of male infertility has focused on the Y chromosome, many other spermatogenesis genes are present throughout the human genome. Recent studies and earlier speculations indicate that the Y chromosome is not the only chromosome that accumulates genes that benefit spermatogenesis over an evolutionary time span (133, 148, 154, 155). Its counterpart, the X chromosome, also seems to be an ideal locus for spermatogenesis genes. Like the Y chromosome, the X chromosome is present as a single copy in the heterogametic XY male. Of course, in the egg, i.e., in the female, the X chromosome undergoes normal meiosis and repair with its paired X mate. However, the X chromosome, like the Y chromosome, does not get to undergo normal meiotic repair in the testis. So it cannot escape completely some of the problems that the Y chromosome faces. Therefore, the X chromosome also has large ampliconic regions. Furthermore, any genetic alteration on the X would have an immediate impact in the male, because there is no other X chromosome to compensate or counteract the effect. Therefore, a recessive gene mutation on the X that is beneficial to the male would preferentially be allocated to the X chromosome as opposed to an autosome. The X chromosome (which is only repaired at meiosis in the egg, not in the testis) has huge ampliconic regions, similarly to the Y chromosome, which have not yet been well characterized and should have a rich complement of spermatogenesis genes.
In fact, reverse-transcription PCR subtraction studies of spermatogonia in mice have demonstrated that a large fraction of genes that are expressed exclusively in premeiotic male germ cells are indeed X chromosomal in origin (155). Eleven of the 36 genes expressed specifically in mouse spermatogonia were found exclusively on the X chromosome. Because the X chromosome is well conserved in all mammals, it seems very likely that evolution has also conferred on the human X chromosome a large portion of the burden for spermatogenesis. (See the Supplemental Material for more).
TRANSMISSION OF Y DELETIONS TO ICSI OFFSPRING
Once it had been shown that many cases of male infertility were caused by deletions on the Y chromosome, concern was registered immediately about the possibility of transmitting these deletions (or other genetic causes of male infertility) to offspring via what was the relatively new technique of ICSI, first introduced in 1992. Although the first boys born from ICSI procedures are not yet 20 years old, it seems likely that if they carry the same genetic defect as their father on their Y chromosome, they will be found to be infertile just as their fathers were.
Microdeletions on the long arm of the Y chromosome do not appear to adversely affect the fertilization or pregnancy results in either severely oligozoospermic or azoospermic men, from whom sperm was successfully retrieved by TESE (3, 9, 10). Concern had been registered that ICSI results might be poorer in Y-deleted men, but this has not been the case (10, 101). Thus, men with Y deletions have the same chance of obtaining offspring via ICSI as non–Y-deleted males undergoing ICSI. Most genes involved in these Y deletions are expressed specifically in the testis during spermatogenesis but do not seem to be essential for fertilization or embryogenesis. The only problem for obtaining a pregnancy is the low number of sperm available in these patients, and this is circumvented by the ICSI procedure.
There has been some concern expressed about a possible widening of the AZFc deletion when it is transmitted to the next generation, but in our centers all of the male offspring from Y-deleted men have had the same Y deletion as their infertile father without any expansion (5, 9, 125, 156). In fact, there was no cogent reason to expect there would be any expansion, considering the mechanism of these Y deletions. So we also examined the Y chromosome of fathers, brothers, and paternal uncles of the infertile men for Y deletions and fertility. In all of the infertile Y-deleted men, the deletions were shown to be completely new, i.e., the fertile fathers of the infertile Y-deleted patients had no Y deletion, the deletion first appearing in the infertile sons. However, all male offspring of these infertile Y-deleted men derived from ICSI procedures had the same Y deletion of their father transmitted to them without any change (Fig. 4) (9). So there is no evidence that the male offspring will be any worse off than his father. Therefore, most couples with male infertility who have a baby are not very troubled by the possibility that their son might have a similar problem (157, 158). (See the Supplemental Materialfor more).
It can therefore be assumed that most Y chromosome deletions arise during spermatogenesis in the fertile testis of the infertile man’s fertile father, and not during embryogenesis. The deletions that arise in the testis of the fertile father are caused by an accidental homologous recombination between large sites of sequence identity causing loss of the intervening sequence. The exact frequency of the occurrence of these deletions in the testis is unknown, although it is estimated that $1 in 1,000 newborn boys are Y-deleted (2). The mechanism for the occurrence of Y deletion in a boy is that 1 out of every 1,000 or so sperm produced in every normal male’s testis has a Y deletion caused by illegitimate homologous recombination. (See the Supplemental Material for more).
CONCLUSIONS ABOUT Y DELETIONS AND ICSI
The use of ICSI has increased tremendously over the past 2 decades and currently allows even azoospermic men with only minute amounts of spermatozoa in their testis to father children. In the majority of cases, there is no obvious explanation for the deficient sperm production in the male other than genetic, and it is possible that male ICSI offspring would inherit the aberration and therefore also be infertile. However, this possibility has rarely deterred the infertile couple from undergoing ICSI. Studies on the role of Y deletions in male infertility have provided new insights into this transmission of male infertility via ICSI and greater understanding of why we can find some few sperm in the testes of azoospermic men. Through evolution, the Y chromosome acquired a specialized role in spermatogenesis, and this has made it highly useful in studying the genetic causes of male infertility. Aberrations on the Y chromosome are currently found in ~15% of severely infertile men. Most, though not all, of these men still possess some degree of spermatogenesis that results in sufficient spermatozoa to perform ICSI and have children. The presence of Y deletions does not decrease the fertilization or pregnancy rate with ICSI, thereby enabling these men to father children with the same efficiency as non–Y-deleted men undergoing ICSI.
It is apparent that there is likely to be frequent transmission of male infertility from the ICSI father to his male offspring, regardless of current Y chromosome testing. Every couple must decide for themselves whether they wish to consider this risk. In our experience, almost all such couples, when well informed, still choose to have ICSI despite this risk (159). Parents usually conclude that if they can have a baby via ICSI, then their male offspring also can.
SUPPLEMENTAL MATERIAL
Genetic Causes of Male Infertility: Role of the Y Chromosome
However, the percentage of male infertility that can be explained by karyotyping alone is low, because of the low resolution of cytogenetic studies. It was not until the development of modern molecular techniques such as polymerase chain reaction (PCR) that the genetic causes of male infertility could be studied with much greater detail. Since then, many more genetic abnormalities, such as microdeletions and point mutations, have been described in infertile males, with most research focusing on the role of genes on the human Y chromosome. Many cases of male infertility are still diagnosed as idiopathic, but they are likely to prove to be genetic in origin also.
AZFa
This finding in the AZFa region runs parallel to our previous observations of spermatogenesis in that larger Y deletions (which take out more genes) are associated with a lesser likelihood of finding sufficient sperm for ICSI than smaller deletions (3, 106).
Evolution and Genetic Constitution of the Human Y Chromosome
As we have explained, massive palindromes, or mirror-image inverted repeats of DNA sequence, characterize the ”ampliconic” portion of the Y chromosome, which contains all the testis-specific genes. In the absence of meiotic pairing, these gene pairs are maintained by intrapalindrome arm-to-arm recombination. This is like the Y chromosome having to pair with itself, because it cannot pair as in normal meiosis to any homologous (or like) chromosome.
But this gene conversion mechanism can result in crossing over between sister chromatids to form instead these isodicentric double strands which just appear to be terminal deletions. This certainly results in spermatogenic failure because of missing distal genes, but there is a double copy of the proximal part of the chromosome. The bigger problem is that the Y chromosome now contains two centromeres, which results in meiotic instability. The farther away the centromeres are from each other, i.e., the longer the isodicentric Y chromosome, the greater will be the chance that the entire chromosome will be deleted, resulting in an XO female Turner embryo, rather than what should have been a sterile male.
The Y Chromosome and Spermatogenesis in Humans and in Apes
The generally accepted reason for this massive discrepancy in spermatogenesis between these three closely related species lies in their differing mating patterns. Chimpanzees congregate in troupes of 30 to 40 in an extended family wherein any female who goes into heat is instantly mounted by every single male in the troupe. Therefore, there is an intense ”competition” among the sperm of the different males for which one will fertilize the eggs of the females. It is far more likely that the male with the highest sperm count, and the biggest testicles, will become the father of the male offspring, because of this high degree of ”sperm competition” in chimpanzees.
In gorillas, it is the opposite. Any female is permanently attached to just one single silverback alpha male, and if she ever gets pregnant, it will have to be with his sperm only. So, in gorillas, there is no sperm competition. That results in small testes with very low sperm counts in these otherwise huge, very macho animals. But why is that? The answer lies in the peculiar instability of the nonrecombining Y chromosome (149).
The multiple nucleotide direct and inverted sequence repeats (amplicons and palindromes) are where all the testis-specific spermatogenesis genes on the Y are located. These areas are prone to frequent deletions caused by nonhomologous, or ”illegitimate” homologous recombination with itself, as already explained, resulting in dropouts of often huge chunks of DNA, causing the concentration of spermatogenesis genes on the Y chromosome to have a very fragile existence. So without sperm competition, sperm counts over eons of time are likely to go down.
Other Male Infertility Genes Not on the Y Chromosome
Future research into genes on the human X chromosome (not just the Y) and their role in the male will provide even more insights into the genetic basis of male infertility. Beyond the X and Y chromosomes, whole-genome sequencing on all of our original study participants may eventually reveal many other testis-specific genes scattered throughout the genome.
Transmission of Y Deletions to ICSI Offspring
This is not the case for the testes of sons dervied from fathers with XXY Klinefelter syndrome. These offspring all have a normal XY karyotype and are likely to be fertile, whereas spermatozoa from AZFc-deleted men carry the same deletion that was originally detected in their somatic cells (156). Thus, it seems that if a patient carries a deletion on the Y chromosome, as determined by analysis of his blood, all of his spermatozoa will have the same deletion, as will all of his sons when these spermatozoa are used for ICSI.
The result of deletions on the Y chromosome is impaired spermatogenesis and not necessarily a failure to father offspring naturally. The degree of impairment, i.e., mild oligozoospermia, severe oligozoospermia, or azoospermia, differs between different deletions and between different patients with the same deletion. Whether these men are able to induce pregnancies depends on the fertility of their sexual partners. The fact that even men with AZFc deletions can, in some instances, father children without the use of assisted reproductiion technologies, illustrates the ability of some wives of men with minute amounts of spermatozoa to conceive naturally (10, 94, 158). Because some deletions, such as partial deletions of the DAZ clusters, potentially have a less dramatic impact on spermatogenesis, these could very well be transmitted naturally more often (126). However, the use of ICSI could contribute considerably to the spreading of these deletions and, as a consequence, reduce the overall fertility within the human population (Fig. 1) (49).
If a genetic aberration that leads to male infertility is located not on the Y, but on an autosome or on the X chromosome, the possible transmission of this aberration is different from transmission of aberrations on the Y chromosome. In contrast to aberrations on the Y chromosome, aberrations on the X chromosome are never transmitted to male offspring, because all boys inherit an X chromosome from their mother only. All girls, however, born from these men with X chromosome aberrations are carriers. Because these women are not affected by the aberration (because it affects only testis-specific genes) and are therefore fertile, they can naturally transmit the aberration to their offspring, giving rise to infertility in the second generation in 50% of boys and causing 50% of girls to be carriers.
REFERENCES
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nature Genet 1995;10:383–93.
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genet 2001;29:279–86.
- Silber SJ, Alagappan R, Brown LG, Page DC. Y chromosome deletions in azoospermic and severely oligozoospermmic men undergoing intracytoplasmic sperm injection after testicular sperm extrac- tion. Hum Reprod 1998;13:3332–7.
- Silber SJ, Repping R. Transmission of male infertility to future generations: lessons from the Y chro- mosome. Hum Reprod 2002;8:217–29.
- Page DC, Silber S, Brown LG. Men with infertility caused by AZFc deletion can produce sons by intracytoplasmic sperm injection, but are likely to transmit the deletion and infertility. Hum Reprod 1999;14:1722–6.
- Saxena R, Brown LG, Hawkins T, Alagappan RK, Skaletsky H, Reeve MP, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nature Genet 1996;14: 292–9.
- Saxena R, de Vries JW, Repping S, Alagappan RK, Skaletsky H, Brown LG, et al. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics 2000;67: 256–67.
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male- specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003;423:825–37.
- Oates RD, Silber S, Brown LG, Page DC. Clinical characterization of 42 oligospermic or azoospermic men with microdeletion of the AZFc region of the Y chromosome, and of 18 children conceived via ICSI. Hum Reprod 2002;11:2813–24.
- Silber SJ, Page DC, Brown LG, Oates R. ICSI results with and without Y chromosomal deletions in men with severe oligozoospermia and azoospermia. 57th Annual Meeting of the ASRM, Orlando, Florida, 2001. Abstract P-83.
- Silber SJ, Rodriguez-Rigau LJ. Quantitative analysis of testicle biopsy: determination of partial obstruction and prediction of sperm count after surgery for obstruction. Fertil Steril 1981;36:480–5.
- Devroey P, Liu J, Nagy Z, Tournaye H, Silber SJ, van Steirteghem AC. Normal fertilization of human oocytes after testicular sperm extraction and intracytoplasmic sperm injection. Fertil Steril 1994;62:639–41.
- Silber SJ, van Steirteghem A, Nagy Z, Liu J, Tournaye H, Devroey P. Normal pregnancies resulting from testicular sperm extraction and intracytoplasmic sperm injection for azoospermia due to maturation arrest. Fertil Steril 1996;66: 110–7.
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science 1997;278: 675–80.
- Lahn BT, Page DC. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat Genet 1999b;21:429–33.
- Lange J, Skaletsky H, van Daalen SKM, Embry SL, Korver CM, Brown LG, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 2009;138:855–69.
- Rodriguez-Rigau LJ, Smith KD, Steinberger E. Relationship of varicocele to sperm output and fertility of male partners in infertile couples. J Urol 1978;120:691–4.
- Nilsson S, Edvinsson A, Nilsson B. Improvement of semen and pregnancy rate after ligation and division of the internal spermatic vein: fact or fiction? Br J Urol 1979;51:591–6.
- Baker HW, Burger HG, de Kretser DM, Lording DW, McGowan P, Rennie GC. Factors affecting the variability of semen analysis results in infertile men. Int J Androl 1981;4:609–22.
- Baker HW, Straffon WG, McGowan MP, Burger HG, de Kretser DM, Hudson B. A controlled trial of the use of erythromycin for men with asthenospermia. Int J Androl 1985;7:383–8.
- Baker HW, Burger HG, de Kretser DM, Hudson B, Rennie GC, Straffon WG. Testicular vein ligation and fertility in men with varicoceles. Br Med J 1985;291:1678–80.
- Schoysman R, Gerris J. Twelve-year follow-up study of pregnancy rates in 1291 couples with idiopathically impathically impaired male fertility. Acta Eur Fertil 1983;14:51–6.
- Baker HW, Kovacs GT. Spontaneous improvement in semen quality: regression toward the mean. Int J Androl 1985;8:421–6.
- Baker HW. Requirements for controlled therapeutic trials in male infertility. Clin Reprod Fertil 1986;4:13–25.
- Vermeulen A, Vandeweghe M, Desylpere JP. Prognosis of subfertility in men with corrected or uncorrected varicocele. J Androl 1986;7:147–55.
- Silber SJ. The relationship of abnormal semen parameters to male fertility. Hum Reprod 1989;4: 947–53.
- Silber SJ. The varicocele dilemma. Hum Reprod Update 2001;7:70–7.
- Nieschlag E, Hertle L, Fischedick A, Behre HM. Treatment of varicocele: counselling as effective as occlusion of the vena spermatica. Hum Reprod 1995;10:347–53.
- Nieschlag E, Hertle L, Fischedick A, Abshagen K, Behre HM. Update on treatment of varicocele: counselling as effective as occlusion of the vena spermatica. Hum Reprod 1998;13:2147–50.
- Devroey P, Vandervorst M, Nagy P, van Steirteghem A. Do we treat the male or his gamete? Hum. Reprod 1998;13:178–85.
- Palermo G, Joris H, Devroey P, van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 1992; 340:17–8.
- van Steirteghem AC, Nagy Z, Joris H, Liu J, Staessen C, Smitz J, et al. High fertilization and implantation rates after intracytoplasmic sperm injection. Hum Reprod 1993;8:1061–6.
- Liu J, Nagy Z, Joris H, Tournaye H, Devroey P, van Steirteghem AC. Intracytoplasmic sperm injection does not require special treatment of the spermatozoa. Hum Reprod 1994;9:112730.
- Liu J, Nagy Z, Joris H, Tournaye H, Smitz J, Camus M, et al. Analysis of 76 total fertilization failure cycles out of 2732 intracytoplasmic sperm injection cycles. Hum Reprod 1995;10:2630–6.
- Nagy ZP, Liu J, Joris H, Verheyen G, Tournaye H, Camus M, et al. The result of intracytoplasmic sperm injection is not related to any of the three basic sperm parameters. Hum Reprod 1995;10: 1123–9.
- Devroey P, Silber S, Nagy Z, Liu J, Tournaye H, Joris H, et al. Ongoing pregnancies and birth after intracytoplasmic sperm injection with frozen-thawed epididymal spermatozoa. Hum Reprod 1995;10:903–6.
- Silber SJ, Nagy ZP, Liu J, Godoy H, Devroey P, van Steirteghem AC. Conventional in-vitro fertilization versus intracytoplasmic sperm injection for patients requiring microsurgical sperm aspiration. Hum Reprod 1994;9:1705–9.
- Silber SJ, van Steirteghem AC, Liu J, Nagy Z, Tournaye H, Devroey P. High fertilization and pregnancy rate after intracytoplasmic sperm injection with spermatozoa obtained from testicle biopsy. Hum Reprod 1995;10:148–52.
- Silber SJ, Nagy Z, Liu J, Tournaye H, Lissens W, Ferec C, et al. The use of epididymal and testicular spermatozoa for intracytoplasmic sperm injection: the genetic implications for male infertility. Hum Reprod 1995;10:2031–43.
- Steinberger E, Tjioe DY. A method for quantitative analysis of human seminiferous epithelium. Fertil Steril 1968;19:959–61.
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev 1972;52:198–236.
- Zukerman Z, Rodriguez-Rigau LJ, Weiss DB, Chowdhury AK, Smith KD. Quantitative analysis of the seminiferous epithelium in human testicular biopsies, and the relation of spermatogenesis to sperm density. Fertil Steril 1978;30:448–55.
- Silber SJ, van Steirteghem AC, Devroey P. Sertoli cell only revisited. Hum Reprod 1995c;10:1031–2.
- Silber SJ, Nagy Z, Devroey P, Tournaye H, van Steirteghem AC. Distribution of spermatogenesis in the testicles of azoospermic men: the presence or absence of spermatids in the testes of men with germinal failure. Hum Reprod 1997;12:2422–8.
- Silber SJ, Nagy Z, Devroey P, Camus M, van Steirteghem AC. The effect of female age and ovar- ian reserve on pregnancy rate in male infertility: treatment of azoospermia with sperm retrieval and intracytoplasmic sperm injection. Hum Reprod 1997;12:2693–700.
- Silber SJ. Microsurgical TESE and the distribution of spermatogenesis in nonobstructive azoospermia. Hum Reprod 2000;15:2278–84.
- Silber SJ. Intracytoplasmic sperm injection today: a personal review. Hum Reprod 1998;13:208–18.
- Silber SJ. The cure and proliferation of male infertility. J Urol 1998b;160:2072–3.
- Faddy MJ, Silber SJ, Gosden RG. Intra-cytoplasmic sperm injection and infertility. Nat Genet 2001;29:131.
- O’Brien SJ, Wildt DE, Bush M. The cheetah in genetic peril. Sci Am 1986;254:84–92.
- O’Brien SJ, Wildt DE, Bush M, Caro TM, FitzGibbon C, Aggundey I, et al. East African chee- tahs: evidence for two population bottlenecks? Proc Natl Acad Sci U S A 1987;84:508–11.
- Short RV. Human reproduction in an evolutionary context. Ann N Y Acad Sci 1995;709:416–25.
- Jacobs PA, Strong JA. A case of human intersexuality having a possible XXY sex-determining mechanism. Nature 1959;183:302–3.
- Kjessler B. Karyotype, meiosis and spermatogenesis in a sample of men attending an infertility clinic. Monogr Hum Genet 1966;2:1–93.
- Olson SD, Magenis RE. Preferential paternal origin of de novo structural rearrangements. In: Daniel A, editor. The cytogenetics of mammalian autosomal rearrangements. New York: Alan R. Liss; 1998. p. 583–99.
- Jacobs PA, Browne C, Gregson N, Joyce C, White H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet 1992;29:103–8.
- Bonduelle M, Legein J, Derde MP, Buysse A, Schietecatte J, Wisanto A, et al. Comparative follow-up study of 130 children born after intracytoplasmic sperm injection and 130 children born after in-vitro fertilization. Hum Reprod 1995;10:3327–31.
- Bonduelle M, Aytoz A. Wilikens. Prospective follow-up study of 1,987 children born after intracytoplasmic sperm injection (ICSI). In: Filicori M, Flamigni C, editors. Treatment of infertility: the new frontiers. Princeton: Communications Media for Education; 1998. p. 445–61.
- Bonduelle M, Aytoz A, van Assche E, Devroey P, Liebaers I, van Steirteghem A. Incidence of chromosomal aberrations in children born after assisted reproduction through intracytoplasmic sperm injection. Hum Reprod 1998;13:781–2.
- Bonduelle M, Camus M, de Vos A, Staessen C, Tournaye H, van Assche E, et al. Seven years of intracytoplasmic sperm injection and follow-up of 1987 subsequent children. Hum Reprod 1999;14:243–64.
- Bonduelle M, Deketelaere V, Liebaers I, Buysse A, Devroey P, van Steirteghem A. Pregnancy outcome after ICSI: a cohort study of 2995 IVF children and 2899 ICSI children. Hum Reprod 2001;16. Abstr. Bk. 1:O-099.
- van Assche E, Bonduelle M, Tournaye H, Joris H, Verheyen G, Devroey P, et al. Cytogenetics of infertile men. Hum Reprod 1996;11:1–24.
- Tuerlings JH, de France HF, Hamers A, Hordijk R, van Hemel JO, Hansson K, et al. Chromosome studies in 1792 males prior to intra-cytoplasmic sperm injection: the Dutch experience. Eur J Hum Genet 1998;6:194–200.
- Egozcue S, Blanco J, Vendrell JM, Garcia F, Veiga A, Aran B, et al. Human male infertility: chromosome anomalies, meiotic disorders, abnormal spermatozoa and recurrent abortion. Hum Reprod Update 2000;6:93–105.
- Chandley A. The chromosomal basis of human infertility. Br Med Bull 1979;35:181–6.
- Tournaye H, Staessen C, Liebars I, van Assche E, Devroey P, Bonduelle M, et al. Testicular sperm recovery in nine 47, XXY Klinefelter patients. Hum Reprod 1996;11:1644–9.
- Schiff JD, Palermo GD, Veeck LL, Goldstein M, Rosenwaks Z, Schlegel PN. Success of testicular sperm extraction [corrected] and intracytoplasmic sperm injection in men with Klinefelter syndrome. J Clin Endocrinol Metab 2005;90: 6263–7.
- Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet 1976;34:119–24.
- Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 1996;5:933–43.
- Reijo R, Alagappan RK, Patrizio P, Page DC. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet 1996;347:1290–3.
- Ma K, Sharkey A, Kirsch S, Vogt P, Keil R, Hargreave TB, et al. Toward the molecular localization of the AZF locus: mapping of microdeletions in azoospermic men within 14 subintervals of interval 6 of the human Y chromosome. Hum Mol Genet 1992;1:29–33.
- Bhasin S, de Kretser DM, Baker HW. Pathophysiology and natural history of male infertility. J Clin Endocrinol Metab 1994;79:1525–9.
- Kent-First MG, Kol S, Muattem A, Ofir R, Manor D, Blazer S, et al. The incidence and possible relevance of Y-linked microdeletions in babies born after intracytoplasmic sperm injection and their infertile fathers. Mol Hum Reprod 1996;2:943–50.
- Kent-First M, Muallem A, Shulz J, Pryor J, Roberts K, Nolten W, et al. Defining regions of the Y-chromosome responsible for male infertility and identification of a fourth AZF region (AZFd) by Y-chromosome microdeletion detection. Mol Reprod Dev 1999;53:27–41.
- Morris RS, Gleicher N. Genetic abnormalities, male infertility, and ICSI. Lancet 1996;347:1277.
- Najmbadi H, Huang V, Yen P, Subbarao MN, Bhasin D, Banaag L, et al. Substantial prevalence of microdeletions of the Y-chromosome in infertile men with idiopathic azoospermia and oligozoospermia detected using a sequence-tagged site-based mapping strategy. J Clin Endocrinol Metab 1996;81:1347–52.
- Nakahori Y, Kuroki Y, Komake R, Kondoh N, Maniki M, Iwamoto T, et al. The Y chromosome region essential for spermatogenesis. Horm Res 1996;46:20–3.
- Prosser J, Inglis JD, Condie A, Ma K, Kerr S, Thakrar R, et al. Degeneracy in human multicopy RBM (YRRM), a candidate spermatogenesis gene. Mamm Genome 1996;7:835–42.
- Qureshi SJ, Ross AR, Ma K, Cooke HJ, Intyre MA, Chandley AC, et al. Polymerase chain reaction screening for Y chromosome microdeletions: a first step toward the diagnosis of genetically-determined spermatogenic failure in men. Mol Hum Reprod 1996;2:775–9.
- Vogt PH, Affara N, Davey P, Hammer M, Jobling MA, Lau YF, et al. Report of the Third International Workshop on Y Chromosome Maping 1997. Cytogenet Cell Genet 1997;79:1–20.
- Elliott DJ, Millar MR, Oghene K, Ross A, Kiesewetter F, Pryor J, et al. Expression of RBM in the nuclei of human germ cells is dependent on a critical region of the Y chromosome long arm. Proc Natl Acad Sci U S A 1997;94:3848–53.
- Elliott DJ, Cooke HJ. Y chromosome microdeletions and male infertility. Hum Fertil 1998;1:64–8.
- Girardi SK, Mielnik A, Schlegel PN. Submicroscopic deletions in the Y chromosome of infertile men. Hum Reprod 1997;2:1635–41.
- Kremer JA, Tuerlings JH, Meuleman EJ, Schoute F, Mariman E, Smeets DF, et al. Microdeletions of the Y chromosome and intracytoplasmic sperm injection: from genet to clinic. Hum Reprod 1997;12: 687–91.
- Kremer JA, Tuerlings JH, Borm G, Hoefsloot LH, Meuleman EJ, Braat DD, et al. Does intracytoplasmic sperm injection lead to a rise in the frequency of microdeletions in the AZFc region of the Y chromosome in future generations? Hum Reprod 1998;13:2808–11.
- Mulhall JP, Reijo R, Alagappan R, Brown L, Page D, Carson R, et al. Azoospermic men with deletion of the DAZ gene cluster are capable of completing spermatogenesis: fertilization, normal embryonic development and pregnancy occur when retrieved testicular spermatozoa are used for intracytoplasmic sperm injection. Hum Reprod 1997;12:503–8.
- Pryor JL, Kent-First M, Muallem A, van Bergen AH, Nolten WE, Meisner L, et al. Microdeletions in the Y chromosome of infertile men. N Engl J Med 1997;336:534–9.
- van der Ven K, Montag M, Peschka B, Leygraaf J, Schwanitz G, Haidl G, et al. Combined cytogenetic and Y chromosome microdeletion screening in males undergoing intracytoplasmic sperm injection. Mol Hum Reprod 1997;3:699–704.
- Vereb M, Agulnik AI, Houston JT, Lipschultz LI, Lamb DJ, Bishop CE. Absence of DAZ gene mutations in cases of nonobstructed azoospermia. Mol Hum Reprod 1997;3:55–9.
- Chai NN, Zhhou H, Hernandez J, Najmabadi H, Bhasin S, Yen PH. Structure and organization of the RBMY genes on the human Y chromosome: transposition and amplification of an ancestral autosomal hnRNPG gene. Genomics 1998;49: 283–9.
- Grimaldi P, Scarponi C, Rossi P, March MR, Fabbri A, Isidori A, et al. Analysis of Yq microdeletions in infertile males by PCR and DNA hybridization techniques. Mol Hum Reprod 1998;4:1116–21.
- Oliva R, Margarit E, Ballesca JL, Carrio A, Sanchez A, Mila M, et al. Prevalence of Y chromosome microdeletions in oligospermic and azoospermic candidates for intracytoplasmic sperm injection. Fertil Steril 1998;70:506–10.
- Stuppia L, Gatta V, Calabrese G, Guanciali FP, Morizio E, Bombieri C, et al. A quarter of men with idiopathic oligo-azoospermia display chromosomal abnormalities and microdeletions of different types in interval 6 of Yq11. Hum Genet 1998;102: 566–70.
- Chang PL, Sauer MV, Brown S. Y chromosome microdeletion in a father and his four infertile sons. Hum Reprod 1999;14:2689–94.
- Kim SW, Kim KD, Paick JS. Microdeletions within the azoospermia factor subregions of the Y chromosome in patients with idiopathic azoospermia. Fertil Steril 1999;72:349–53.
- Krausz C, Bussani-Mastellone C, Granchi S, McElreavey K, Scarselli G, Forti G. Screening for microdeletions of Y chromosome genes in patients undergoing intracytoplasmic sperm injection. Hum Reprod 1999;14:1717–21.
- Seifer I, Amat S, Delgado-Viscogliosi P, Boucher D, Bignon YJ. Screening for microdeletions on the long arm of chromosome Y in 53 infertile men. Int J Androl 1999;22:148–54.
- Cram DS, Ma K, Bhasin S, Arias J, Pandjaitan M, Chu B, et al. Y chromosome analysis of infertile men and their sons conceived through intracytoplasmic sperm injection: vertical transmission of deletions and rarity of de novo deletions. Fertil Steril 2000;74:909–15.
- van Landuyt L, Lissens W, Stouffs K, Tournaye H, Liebaers I, van Steirteghem A. Validation of a simple Yq deletion screening programme in an ICSI candidate population. Mol Hum Reprod 2000;6:291–7.
- Krausz C, McElreavey K. Y chromosome microdeletions in ”fertile” males. Hum Reprod 2001;16: 1306–7.
- van Golde RJ, Wetzels AM, de Graaf R, Tuerlings JH, Braat DD, Kremer JA. Decreased fertilization rate and embryo quality after ICSI in oligozoospermic men with microdeletions in the Azoospermia Factor c region of the Y chromosome. Hum Reprod 2001;16:289–92.
- Brown GM, Furlong RA, Sargent CA, Erickson RP, Longepied G, Mitchell M, et al. Characterisation of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to theSxrb interval of the mouse Y chromosome of the Dffry gene. Hum Mol Genet 1998;7:97–107.
- Sargent CA, Boucher CA, Kirsch S, Brown G, Weiss B, Trundley A, et al. The critical region of overlap definings theAZFa male infertility interval of proximal Yq contains three transcribed sequences. J Med Genet 1999;36:670–7.
- Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, et al. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nature Genet 1999;23:429–32.
- Sun C, Skaletsky H, Rozen S, Gromoll J, Niesclag E, Oates R, et al. Deletion of Azoospermia Factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet 2000;9:2291–6.
- Repping S, Skaletsky H, Lange J, Silber S, van der Veen F, Oates RD, et al. Recombination between palindromes P5 to P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet 2002;71:906–22.
- Brandell RA, Mielnik A, Liotta D, Ye Z, Veeck LL, Palermo GD, et al. AZFb deletions predict the absence of spermatozoa with testicular sperm extraction: preliminary report of a prognostic genetic test. Hum Reprod 1998;13:2812–5.
- Martinez MC, Bernabe MJ, Gomez E, Ballesteros A, Landers J, Glover G, et al. Screening for AZF deletion in a large series of severely impaired spermatogenesis patients. J Androl 2000;21:651–5.
- Ma K, Inglis JD, Sharkey A, Bickmore WA, Hill RE, Prosser EJ, et al. A Y chromosome gene family with RNA-binding protein homology: candidates for the azoospermia factor AZF controlling human spermatogenesis. Cell 1993;75:1287–95.
- Foote S, Vollrath D, Hilton A, Page DC. The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science 1992;258:60–6.
- Kobayashi K, Mizuno K, Hida A, Komaki R, Tomita K, Matsushita I, et al. PCR analysis of the Y chromosome long arm in azoospermic patient: evidence for a second locus required for spermatogenesis. Hum Mol Genet 1994;3:1965–7.
- Chai NN, Salido EC, Yen PH. Multiple functional copies of the RBM gene family, a spermatogenesis candidate on the human Y chromosome. Genomics 1997;45:355–61.
- Affara N, Bishop C, Brown W, Cooke H, Davey P, Ellis N, et al. Report of the Second International Workshop on Y Chromosome Mapping 1995. Cyto- genet Cell Genet 1996;73:33–76.
- Menke DB, Mutter GI, Page DC. Expression of DAZ, an azoospermia factor candidate, in human spermatogonia. Am J Hum Genet 1997;60:237–41.
- Hackstein JH, Hochstenbach R. The elusive fertility genes of Drosophila: the ultimate haven for selfish genetic elements. Trends Genet, 11, 195–200
- Cooke HJ, Lee M, Kerr S, Ruggiu M. A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet 1996;5:513–6.
- Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 1996;381:783–5.
- Karashima T, Sugimoto A, Yamamoto MA. A C. elegans homologue of DAZ/boule is involved in progression through meiosis during oogenesis. Worm Breeder’s Gaz 1997;15:65–6.
- Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997;389:73–7.
- Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development 1998;125:171–80.
- Xu EY, Moore FL, Pera RA. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci U S A 2001;98:7414–9.
- Saut N, Terriou P, Navarro A, Levy N, Mitchell MJ. The human Y chromosome genes BPY2, CDY1 and DAZ are not essential for sustained fertility. Mol Hum Reprod 2000;6:789–93.
- Moro E, Ferlin A, Yen PH, Franchi PG, Palka G, Foresta C. Male infertility caused by a de novo partial deletion of theDAZ clusters on the Y chromosome. J Clin Endocrinol Metab 2000;85:4069–73.
- Bienvenu T, Patrat C, McElreavey K, de Almeida M, Jouannet P. Reduction in the DAZ gene copy number in two infertile men with impaired spermatogenesis. Ann Genet 2001;44:125–8.
- de Vries JW, Hoffer MJ, Repping S, Hoovers JM, Leschot NJ, van der Veen F. Reduced copy number of DAZ genes in subfertile and infertile men. Fertil Steril 2002;77:68–75.
- Repping S, van Daalen SK, Korver CM, Brown LG, Marsyalek JD, Gianolten J, et al. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8Mb deletion in AZFc region. Genomics 2004;83:1046–52.
- Blanco P, Shlumukova M, Sargent CA, Jobling MA, Affara N, Hurles ME. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J Med Genet 2000;37:752–8.
- Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet 2000;9:2563–72.
- Rozen S, Skaletsky H, Marsyalek JD, Minx PJ, Cordum HS, Waterston RH, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosome. Nature 2003;423:873–6.
- Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature 1998;394:776–80.
- Jegalian K, Lahn BT. Why the Y is so weird. Sci Am 2001;284:56–61.
- Rice WR. Sexually antagonistic genes: experimental evidence. Science 1992;256:1436–9.
- Rice WR. Degeneration of a nonrecombining chromosome. Science 1994;263:230–2.
- Rice WR. Evolution of the Y sex chromosome in animals. BioScience 1996;46:331–43.
- Graves JA. The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. Bioessays 1995;17:311–20.
- Graves JA. The evolution of mammalian sex chro- mosomes and the origin of sex determining genes. Philos Trans R Soc Lond B Biol Sci 1995;350: 305–11.
- Graves JA, Disteche CM, Toder R. Gene dosage in the evolution and function of mammalian sex chromosomes. Cytogenet Cell Genet 1998;80:94–103.
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science 1999;286:964–7.
- Delbridge ML, Harry JL, Toder R, O’Neill RJ, Ma K, Chandley AC, et al. A human candidate spermatogenesis gene, RBM1, is conserved and amplified on the marsupial Y chromosome. Nature Genet 1997;15:131–6.
- Delbridge ML, Lingenfelter PA, Disteche CM, Graves JA. The candidate spermatogenesis gene RBMY has a homologue on the human X chromosome. Nature Genet 1999;22:223–4.
- Mazeyrat S, Saut N, Mattei MG, Mitchell MJ. RBMY evolved on the Y chromosome from a ubiquitously transcribed X-Y identical gene. Nat Genet 1000:22:224–6.
- Vogel T, Speed RM, Teague P, Cooke HJ. Mice with Y chromosome deletion and reduced Rbm genes on a heterozygous Dazl1 null background mimic a human azoospermic factor phenotype. Hum Reprod 1999;14:3023–9.
- Graves JA. Two uses for old SOX. Nat Genet 1997;16:114–5.
- Pask A, Graves JA. Sex chromosomes and sex-determining genes: insights from marsupials and nomotremes. Cell Mol Life Ser 1999;55:71–95.
- Vidal VP, Chaboissier MC, de Rooij DG, Schell A. Sox9 induces testis development in XX transgenic mice. Nature Genet 2001;28:216–7.
- Winge O. The location of eighteen genes in lebistes reticulates. J Genet 1927;18:1–43.
- Fisher RA. The evolution of dominance. Biol Rev 1931;6:345–68.
- Charlesworth D, Charlesworth B. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet Res 1980;35:205–14.
- Silber SJ. The disappearing male. In: Jansen R, Mortimer D, editors. Toward reproductive certainty— fertility and genetics beyond 1999. New York/London: Parthenon Publishing Group; 1999. p. 499–505.
- Brooks R. Negative genetic correlation between male sexual attractiveness and survival. Nature 2000;406:67–70.
- Enomoto T, Matsubayashi K, Nakano M, Fuji-Hanamoto H, Kusunoki. Testicular histological examination of spermatogenic activity in captive gorillas. Am J Primatol 2004;63:183–99.
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SKM, Saskiak M, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 2010;463:536–9.
- Noordam MJ, van Daalen SKM, Hovingh SE, Korver CM, van der Veen F, Repping S. A novel partial deletion of the Y chromosome azoospermia factor C region is caused by nonhomologous recombination between palindromes and may be associated with increased sperm counts. Hum Reprod 2011;26:713–23.
- Rice WR. Sex-chromosomes and the evolution of sexual dimorphism. Evolution 1984;38:735–42.
- Wang PG, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 2001;27:422–6.
- de Vries JW, Repping S, Oates R, Carson R, Leschot NJ, van der Veen F. Absence of Deleted in Azoospermia (DAZ) genes in spermatozoa of infertile men with somatic DAZ deletions. Fertil Steril 2001;75:476–9.
- Edwards RG, Bishop CE. On the origin and frequency of Y chromosome deletions responsible for severe male infertility. Mol Hum Reprod 1997;3: 549–54.
- Sokol RZ, Sparkes R. Demonstrated paternity in spite of severe idiopathic oligospermia. Fertil Steril 1987;47:356–8.
- Giltay JC, Kastrop PM, Tuerlings JH, Kremer JA, Tiemessen CH, Gerssen-Schoorl KB, et al. Subfertile men with constitutive chromosome abnormalities do not necessarily refrain from intracytoplasmic sperm injection treatment: a follow-up study on 75 Dutch patients. Hum Reprod 1999;14:318–20.

