Our Research on The Y Chromosome and Male Infertility
Along with the development of ICSI in 1993, our center was the first to study the Y chromosome and male infertility, and why tiny amounts of sperm are often found in the testes of azoospermic men previously thought to be making no sperm. You have probably heard a lot about the Y chromosome. It is what determines that a male is a male. We discovered with our first scientific paper on this in 1995 that the Y chromosome contains many genes that are involved in spermatogenesis, and deletions involving these genes are often found in infertile males. There has been a great deal of unknowledgeable discussion about our discovery of these sperm producing genes on the Y and a lot of misinformation. So in this page, I will try to clear up the confusion so you will understand better the genetics of male infertility. Our sequencing the DNA of the Y gives us a deep perspective about infertility genes that are widespread throughout the genome and which are also involved in transmitting infertility to future generations. A benefit of understanding the Y chromosome is it will help us to comprehend why men who are seemingly azoospermic usually have some residual tiny amount of spermatogenesis that can be used for successful ICSI. More importantly, it will expose the futility of trying to increase sperm count with drugs or varicocoele surgery.
Until two decades ago, there were no treatment options for infertile couples when the male had severely impaired spermatogenesis. In fact, there are still no clinical therapies to correct deficient spermatogenesis. Since the introduction of ICSI by us and the Brussels Dutch-Speaking Free University in 1992, however, there has been a revolution in our thinking about male infertility. Infertile couples with the most severe cases of male infertility, even with apparently 100% abnormal morphology and even just rare spermatozoa in the ejaculate, can now have pregnancy and delivery rates not apparently different from conventional IVF with normal sperm.
In 1993, we were the first to introduce microsurgical epididymal sperm aspiration (MESA) in conjunction with ICSI for the treatment of obstructive azoospermia. A few months later, TESE (testicular sperm extraction) was also found to be effective for the majority of cases of non-obstructive azoospermia as well. The reason is that approximately 60% of azoospermic men with presumably no sperm production actually do have a minute amount of sperm production in the testis that is just not quantitatively sufficient to spill over into the ejaculate, but which is adequate for ICSI. Thus, even men with spermatogenesis so deficient in quantity that no sperm at all can reach the ejaculate, could now have children with the use of TESE-ICSI.
Infertile couples from all over the world come to St. Louis, Missouri, to chase their dream, because Dr. Sherman Silber and his team are simply the best there is.
Discovery Health Channel Documentary

The Y chromosome and spermatogenesis in humans and in apes
Comparing spermatogenesis in humans, chimpanzees and gorillas has always been fascinating. Chimpanzees, which weigh only about 100 pounds have enormous 8 centimeter diameter round (not oval) testes with sperm counts of over a billion per ml. Yet gorillas, which weight as much as 600 pounds or more, have tiny testes, very poor spermatogenesis, and in the sparse literature on gorilla testicular histology, in the majority of cases have what appears to be Sertoli cell only. Humans, the closest living relatives to chimpanzees and gorillas, fall somewhere in between.
The generally accepted reason for this massive discrepancy in spermatogenesis between these three closely related species lies in their differing mating patterns. Chimpanzees congregate in troupes of 30 to 40 in an extended family wherein any female who goes into heat is instantly mounted by every single male in the troupe. Therefore, there is an intense “competition” between the sperm of the different males to see which one will fertilize the females’ eggs. It is far more likely that the male with the highest sperm count, and the biggest testicles, will become the father of the male offspring, because of the high degree of “sperm competition” in chimpanzees.
In gorillas, it is the opposite. Any female is permanently attached to just one single silverback alpha male, and if she ever gets pregnant, it will have to be with his sperm only. So in gorillas there is no sperm competition. That results in small testes with very low sperm counts in these otherwise huge, very macho animals. But why is that? The answer lies in the peculiar instability of the non-recombining Y chromosome.
The multiple nucleotide sequence direct and inverted repeats (amplicons and palindromes) are where all the testis specific spermatogenesis genes on the Y are located. These areas are prone to frequent deletions caused by non-homologous, or “illegitimate” homologous recombination with itself, resulting in drop-outs of often huge chunks of DNA, making the concentration of spermatogenesis genes on the Y chromosome have a very fragile existence. So without sperm competition, sperm counts over eons of time are likely to go down.
Most intriguing is to compare the human Y chromosome to the chimpanzee Y chromosome, both of which have been fully and accurately sequenced. Unfortunately, the gorilla Y has not yet been sequenced. Nonetheless some interesting differences are noted between the human Y and the chimpanzee Y. Firstly, the chimpanzee Y has many more amplicons and palindromes than the human, and nonetheless, much fewer ampliconic genes (25 compared to 60). Furthermore, the chimpanzee Y is missing a gene (PRY) that is present on AZFc in the human, but completely absent in the chimpanzee. Furthermore, this gene has also been found to be absent in rare humans who have incredibly high sperm counts, approaching half a billion. Thus, comparing the super fertile chimpanzee Y chromosome to its less fertile human cousin, can help us understand better the genetic control of spermatogenesis in our infertile male patients.
Genetic causes of male infertility: role of the Y chromosome
Chromosomal abnormalities in males, such as translocations and inversions, which can be found with routine karyotyping, are found in approximately 1% of azoospermic males. However, the most common chromosomal abnormalities in azoospermic men are abnormalities involving the sex chromosomes, which are found in approximately 4% of these men. Klinefelter’s syndrome, in which patients show a 47,XXY karyotype, is the most frequent form. Even in these cases, we can usually find a few rare sperm adequate for fertility using ICSI.
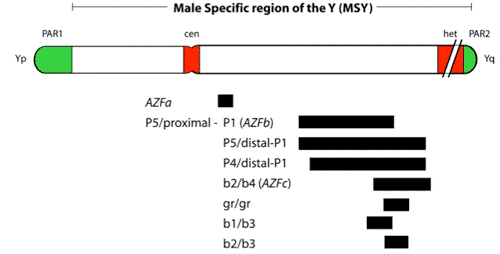
However, the percentage of male infertility that can be explained by karyotyping alone is low, this being mainly due to the low resolution of routine cytogenetic studies. It was not until the development of modern molecular techniques such as polymerase chain reaction (PCR) that we could study the genetic causes of male infertility with much greater detail. Since then, many more genetic abnormalities, such as micro-deletions and point mutations, have been described in infertile males, with most research focusing on the role of genes on the human Y chromosome. We have shown that the long arm of the Y chromosome contains not one but many distinct deletion intervals and at least 60 genes belonging to nine gene families whose exclusive function is in spermatogenesis.
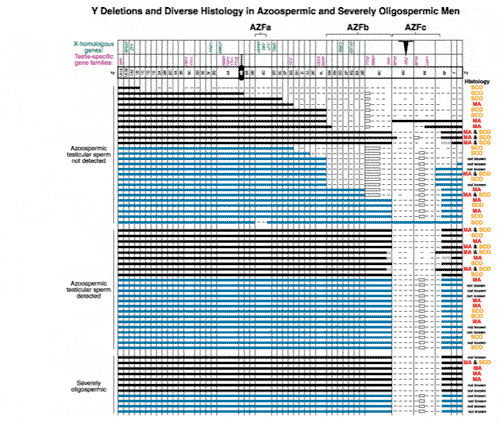
In fact, the deletion frequency of one or more of these regions on the Y chromosome in men with azoospermia or severe oligozoospermia is approximately 15% (Figure 1). After our initial report, many laboratories throughout the world have reported on these sub-microscopic deletions of the Y chromosome in azoospermic and severely oligozoospermic men. In fact, deletion screening of the Y chromosome is now considered standard practice for severely oligozoospermic and azoospermic patients undergoing assisted reproduction in most countries in the world.
Chimpanzees have sperm competition in their mating pattern. Chimpanzee testicles and sperm production are huge because of its better Y chromosome. Gorillas have no sperm competition in their mating system. Gorillas have small testicles and very poor sperm production because lack of sperm competition causes deteriation of the Y chromosome.
AZFa & b
The AZFa region differs from the AZFb and AZFc regions because of its non-repetitive structure and its low deletion frequency. AZFa deletions are very very uncommon and only a few rare patients have been described (Figure 2). However, studies of this region are highly useful in understanding the genetic basis of male infertility. The lack of sequence repeats which plague the rest of the Y chromosome, made this particular region of the Y amenable to a point mutation search. Because almost all other spermatogenesis genes on the Y chromosome, such as those in AZFb and AZFc, are multicopy, searching for point mutations in these genes is virtually impossible. It is also questionable whether point mutations in a single gene of a gene family will give rise to a severe infertility phenotype as the remaining intact copies of the gene could potentially compensate for the loss of function of the mutated gene. This is an important lesson for why spermatogenic defects come in varying degrees of severity, and why even in azoospermic men, there are usually still a tiny number of healthy sperm “hiding” in the testicles.
The AZFa region thus also provides a good model for the interaction and overlapping functions of multiple genes which sheds light on the polygenic nature of the genetic control of spermatogenesis. Larger Y deletions (which take out more genes) are associated with a lesser likelihood of finding sufficient sperm for ICSI than smaller deletions (Figure 3).
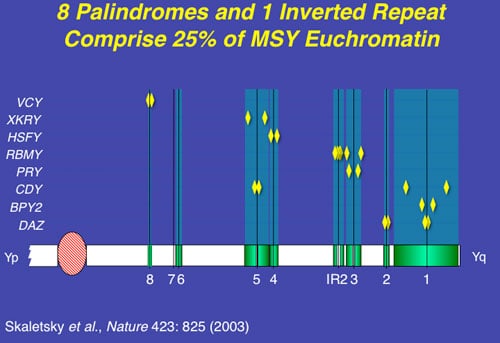
Deletions of the AZFb region are slightly more common than deletions of the AZFa region but are still found in a very small percent of azoospermic men. Interestingly, all men with deletions of AZFb described to date are azoospermic and also show complete absence of spermatozoa in the testis. In fact, the region frequently referred to simplistically as AZFb overlaps AZFc and has a massive number of repeat copies of genes and pseudogenes (RBMY, PRY, TTTY) arranged in a complex of palindromes (inverted repeat sequences). Deletions in this region of the Y are massive and are much less common than in AZFc, and are caused by weak “breakpoints” in the center of its inverted repeats.
AZFc

AZFc is the first region of the Y we studied completely in fertile and infertile men. The most commonly deleted and best-studied region on the Y chromosome is the AZFc region. Deletion of the AZFc region is found in approximately 12% of azoospermic men and in 6% of severely oligozoospermic mens. The complete nucleotide sequence of the AZFc region revealed an extraordinary structure and genetic composition. The region is constructed from massive areas of absolute sequence identity called amplicons which are arranged in direct repeats, and inverted repeats or palindromes. The AZFc region spans 3.5 Mb and contains seven separate families of genes with a total of 19 genes that are all exclusively expressed in the testis (Figure 3). Interestingly, absence of this large 3.5-Mb AZFc chunk of the Y chromosome seems to have no other deleterious effect on the male except upon spermatogenesis exemplifying the remarkably specialized function of this region of the Y. These genes only affect spermatogenesis and nothing else.
The DAZ gene family, which is one of the seven gene families located within AZFc, was one of the first spermatogenesis genes identified on the human Y chromosome. The human DAZ genes were shown to be transcribed specifically in spermatogonia and in early primary spermatocytes. Interestingly, homologues of DAZ in other species were also shown to be involved in control of spermatogenesis, supporting and essential role of this gene in male fertility in humans as well as almost all other animals. Homologues of DAZ have been found in Drosophila (termed Boule), in mice (termed Dazl), in frogs (termed Xdazl) and even in worms (termed daz-1). Therefore, DAZ is the most ancient and well conserved spermatogenic gene. In contrast to humans, the DAZ gene in these other species is single copy and located on an autosome rather than on the Y chromosome. In the human, DAZ is present on the Y in four near-identical repeat copies (99.9% homology) arranged in two clusters with two genes in each cluster.
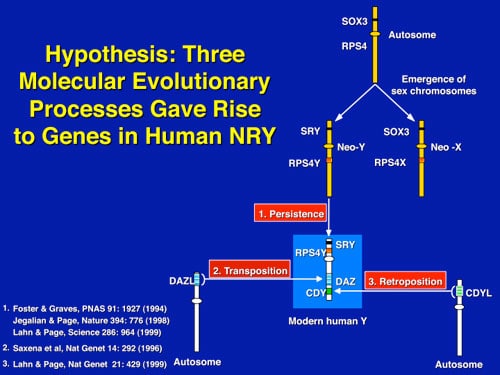
The human also retains an autosomal homologue of DAZ called DAZL, which is located on chromosome 3s. During evolution, some time after the split of Old and New World monkeys approximately 30 million years ago, the DAZL gene was transposed to the Y chromosome (Figure 4). Once autosomal DAZL was transposed to the Y, it was amplified and pruned until it became the modern-day DAZ gene family of four on the Y. In fact there is yet another DAZ family homologue in humans, termed BOULE, that resembles the fly homologue boule, even more closely than DAZ or DAZL. The exact interaction and possible functional overlap between these three members of this interesting gene family holds the clue to the peculiar finding a few surviving sperm in the testes of azoospermic men.
It is this multiplicity of genes copies all of which contribute quantitatively to total sperm production that explains the minute amount of sperm we can find at TESE in azoospermic men, because of “rescue” from the continued presence of at least one of these gene copies even though most are missing or deleted.
Deletions of the entire AZFc region result in loss of all four DAZ copies. Deletions involving only some of the DAZ genes are also found in infertile males, but these smaller deletions are found in men with only mild oligozoospermia, indicating a possible gene dosage effect, i.e. men with a deletion of only two DAZ genes are less affected than men with a deletion of all four copies. These data illustrate that infertility is a complex multigenic disorder and that disruption of different genes or disruption of some genes of a gene family can result in different degrees of spermatogenic failure.
Evolution and genetic constitution of the human Y chromosome
What makes the Y chromosome, with its confounding repeats, polymorphisms, and degenerating regions, such an interesting study object for male infertility? The answer lies in the evolutionary history of the X and Y chromosomes. Over the course of the past 240-320 million years of mammalian evolution, the X and Y chromosomes have evolved from what was originally a pair of ordinary autosomes (Figure 5). During that evolution, just as most of the ancestral X genes were decaying on the Y because of the lack of meiotic recombination, genes which control spermatogenesis arrived on the Y from autosomes. Once on the Y, these formerly autosomal genes amplified into multiple copies, and achieved greater prominence through a process called “gene conversion.” Spermatogenesis genes that arrived on the Y, but came originally from autosomes, include the DAZ (from autosome 3) and CDY (from autosome 6) genes that are among the seven gene families located in AZFc (Figure 6). Other spermatogenesis genes on the Y, such as RBMY, have persisted in their original position as on the X. The ancestral gene that remained on the X chromosome (RBMX) retained its widespread cellular functions, whereas RBMY, which persisted on the receding Y chromosome, evolved a male-specific function in spermatogenesis. Male benefit genes have thus arrived and accumulated on the evolving Y chromosome over many millions of years.
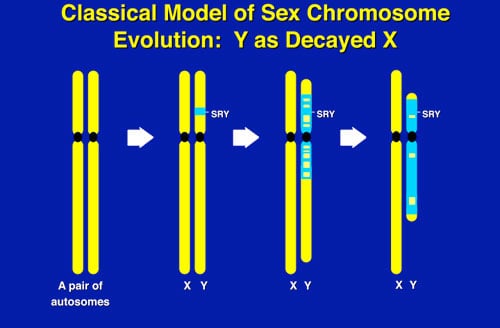
This evolution of the modern X and Y chromosomes was initiated by the emergence of a male sex-determining gene (now known as SRY) on what was originally an ordinary pair of autosomes (Figure 5). Genes associated with the non-recombining SRY region that were specifically beneficial for male function or antagonistic to female function, flourished on the evolving Y chromosome despite the deterioration of more generalized genes which lacked the DNA repair benefits of meiosis.
The next question that the evolution of the X and Y chromosomes presents, which is crucial for understanding male infertility and ICSI is how did this degraded Y chromosome survive at all? The lack of recombination of the male determining Y chromosome has led to complete deterioration and loss of most of its original 1,438 genes (the number of genes on its corresponding X) and an accumulation of only 60 genes (only 9 gene families) that are male specific which are located in areas of sequence identity that promotes further deletions. So how does the Y survive at all, and why do we humans retain any spermatogenesis at all?
The answer is “gene conversion.” It answers the question of how these ampliconic repeats and palindromic inversions occur at all. When autosomes recombine during meiosis, DNA is exchanged in a way that the accumulated mutational errors of life get corrected in the germ cells via this DNA exchange. In a sense, the autosomes have “sex” with each other. This correctional meiosis cannot occur with the Y chromosome. Instead, the Y has “sex” with itself. That is, the like sequence repeats of the Y just “recombine” in a sense with each other, “illegitimate” homologous recombination. This “gene conversion” creates and repairs these multiple copies and inverted DNA sequence repeats that characterize the Y and indeed all sex determining chromosomes. So if there is a missing or deleted copy of a spermatogenesis gene, there are other backup copies that can still rescue spermatogenesis to some degree.
Conclusions about Y deletions and ICSI
The use of ICSI has increased tremendously over the past decade and currently even allows azoospermic men with only minute amounts of spermatozoa in their testis to father children. In the majority of cases, there is no obvious explanation for the deficient sperm production in the male other than genetic, and it is conceivable that male ICSI-offspring will inherit the aberration and thus also be infertile. This possibility will rarely deter the infertile couple from undergoing ICSI. Studies on the role of Y deletions in male infertility have provided new insights into why we can find sperm in azoospermic men. Aberrations on the Y chromosome are currently found in approximately 15% of infertile males. Most, though not all, of these males still possess some degree of spermatogenesis that results in sufficient spermatozoa to perform ICSI and have children. The presence of Y deletions does not decrease the fertilization or pregnancy rate, thereby enabling these men to father children with the same efficiency as non-Y-deleted men undergoing ICSI.
If you have any questions, you may call us at (314) 576-1400.