By: Erica C. Pandolfi, Ernesto J. Rojas, Enrique Sosa, Joanna J. Gell, Timothy J. Hunt, Sierra Goldsmith, Yuting Fan, Sherman J. Silber Amander T. Clark
Abstract
We generated three human induced pluripotent stem cell (hiPSC) sublines from human dermal fibroblasts (HDFs) (MZT04) generated from a skin biopsy donated from a previously fertile woman. The skin biopsy was broadly consented for generating hiPSC lines for biomedical research, including unique consent specifically for studying human fertility, infertility and germ cells. hiPSCs were reprogrammed using Sendai virus vectors and were subsequently positive for markers of self-renewal including OCT4, NANOG, TRA-1-81 and SSEA-4. Pluripotency was further verified using teratomas and PluriTest. These sublines serve as controls for hiPSC research projects aimed at understanding the cell and molecular regulation of female fertility and infertility.
Resource utility
Infertility is a condition that affects approximately 12% of the world’s reproductive age population. Here, we generated control hiPSC sublines that can be responsibly and ethically used for human fertility and infertility research.
Resource details
The use of germ cells and gametes derived from hiPSCs for research on fertility and causes of infertility can raise objections due to the moral significance of reproduction. Strong opinions on the implications of reproductive research and potential applications, leads to the necessity to inform donors that the research specifically allows for reproductive science research to create germ cells and gametes from hiPSC derivatives (Aalto-Setälä et al., 2009). In addition to this specific consent, we believe that it is necessary to receive broader consent from donating patients that allows for future unanticipated research to facilitate the sharing of generated hiPSC lines and sublines. Thus, we created hiPSC sublines MZT04D, MZT04J, and MZT04C to fill a current void in the availability of control hiPSCs for fertility and infertility research.
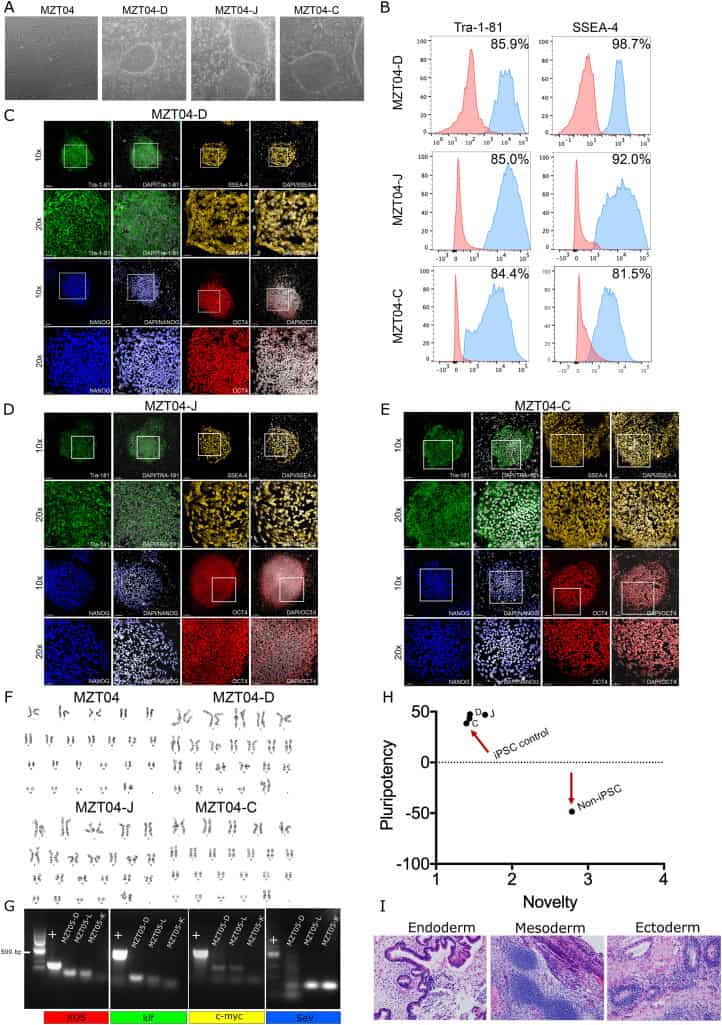
We generated three integration-free hiPSC sublines from human dermal fibroblasts (HDFs). Fibroblasts were derived from a skin punch biopsy from a 55-year-old woman who was fertile prior to entering menopause. These fibroblasts (MZT04) were reprogrammed to hiPSCs using the non-integrating recombinant Sendai virus containing reprogramming factors OCT3/4, SOX2, KLF4 and c-MYC4 (Tokusumi et al., 2002). Twenty-seven days after the transduction, individual colonies were manually picked onto mouse embryonic fibroblast feeder cells to create the sublines. We selected three sublines called MZT04D and MZT04J and MZT04C and characterized them for their pluripotency potential (Table 1). All hiPSC sublines exhibited typical pluripotent stem cell morphology (Fig. 1A) and markers of self-renewal, as confirmed through flow cytometry (Fig. 1B) and Immunofluorescence staining for NANOG, OCT4, and TRA-1-81 and SSEA-4 (Fig. 1C–E). The reprogrammed cells and the initial fibroblasts displayed a normal 46, XX karyotype (Fig. 1F), and did not express the exogenous reprogramming factors after continued culture (Fig. 1G) (expected band size SeV: 181 bp, c-MYC: 532 bp, klf: 410 bp, KOS: 501 bp). To determine the pluripotency of these lines, MZT04D, MZT04J, MZT04C were assessed with a PluriTest analysis (Müller et al., 2011) (Fig. 1H). Differentiation potential was also assessed using teratoma formation assays. Representative of the three germ-layers included for MZT04D (Fig. 1I). To confirm that the hiPSC sublines were of the same genetic background as the donated fibroblasts, short tandem repeat (STR) analysis was conducted demonstrating that each of the three hiPSC lines were identical to the original HDFs (MZT04). We also confirmed that MZT04D, MZT04J, and MZT04C were negative for mycoplasma through routine mycoplasma testing (Supplementary Fig. 1).
Table 1. Summary of lines.
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| UCLAi001-A | MZT04-D | Female | 55 | unknown | N/A | None |
| UCLAi001-B | MZT04-J | Female | 55 | unknown | N/A | None |
| UCLAi001-C | MZT04-C | Female | 55 | unknown | N/A | None |
Materials and methods
Fibroblast derivation
A 1 mm skin punch biopsy was dissected and then digested in Collagenase IV (Life Technologies) for 1 h at 37°, 5.0% CO2. The digested pieces were then plated down on 0.1% gelatin (Sigma) coated (Millipore) plates in human fibroblast media, 15% Fetal bovine serum (GE Healthcare), 1% Non-Essential Amino Acids (Invitrogen), 1% Glutamax, (GibcoTM), 1% Penicillin-Strepromyocin-Glutamine (Gibco), and Primocin (Invivogen), at 37°, 5.0% CO2. Outgrowths of fibroblasts were monitored for two weeks and the media was changed every three days. Fibroblasts were passaged using 0.05% Tryspin (Gibco) and re-plated, the derived cells were termed MZT04 (Tables 2 and 3).
Table 2. Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Phase Contrast | Normal | Fig. 1 panel A |
| Phenotype | Immunofluorescence | Positive for self-renewal markers: Oct4, Nanog, SSEA-4, Tra-1-81 | Fig. 1 panel C-E |
| Flow cytometry | MZT04-D: Tra 1-81: 85.9%, SSEA-4: 98.7% | Fig. 1 panel B | |
| MZT04-J: Tra 1-81: 92.7%, SSEA-4: 81.5% | |||
| MZT04-C: Tra 1-81: 84.3%, SSEA-4: 81.5% | |||
| Genotype | Karyotype (G-banding) and resolution | 46,XX | Fig. 1 panel F |
| Identity | Microsatellite PCR (mPCR) ORSTR analysis | Performed | Supplementary Fig. 1 |
| 16 sites tested, all three lines match each other, and the HDF line they were derived from | Supplementary Fig. 1 | ||
| Mutation analysis (IF APPLICABLE) | Sequencing | N/A | |
| Southern Blot OR WGS | N/A | ||
| Microbiology and virology | Mycoplasma | Mycoplasma testing by Luminescence | Supplementary Fig. 1 |
| Differentiation potential | Teratoma formation and PluriTest | Proof of three germlayers formation | Fig. 1 panel H Fig. 1 panel I |
| Donor screening (OPTIONAL) | N/A | ||
| Genotype additional info (OPTIONAL) | N/A | ||
| N/A |
Table 3. Reagents details.
| Antibodies used for immunocytochemistry/flow-citometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Self-renewal markers | goat-anti-human Oct4 | 1:100 | Santa Cruz, sc8628 |
| RRID: AB_653551 | |||
| Self-renewal markers | goat-anti-human NANOG | 1:40 | R&D Systems, AF1997 |
| RRID: AB_355097 | |||
| Self-renewal markers | mouse-anti-human SSEA-4 | 1:100 | Developmental Studies Hybridoma |
| Bank, MC-813-70 | |||
| RRID: AB_528477 | |||
| Self-renewal markers | mouse-anti-human TRA-1-81 | 1:100 | eBiosciences, 14-8883-82 |
| RRID: AB_891614 | |||
| Pluripotency markers | SSEA-4-Allophycocyanin | 1:30 | R&D Systems, FAB1435A |
| RRID: AB_494994 | |||
| Pluripotency markers | TRA-1-85-Phycoerythrin | 1:60 | R&D Systems, FAB3195P |
| RRID: AB_2066683 | |||
| Pluripotency markers | TRA-1-81, Alexa Fluor 488 | 1:60 | Stemcell Technologies, |
| 60065AD | |||
| RRID: AB_2721032 | |||
| Pluripotency markers | Dapi | 1:100 | BioVision, B1098-25 |
| RRID: AB_2336790 | |||
| Secondary antibodies | AF488-conjugated donkey-anti-goat | 1:200 | JacksonImmunoResearch, 705- |
| 546-147 | |||
| RRID: AB_2340430 | |||
| Secondary antibodies | AF488-conjugated donkey-anti-mouse | 1:200 | Life Technologies, A-21131 |
| RRID: AB_2535771 |
| Primers | ||
|---|---|---|
| Target | Forward/Reverse primer (5′–3′) | |
| Reprogramming virus | SeV | GGA TCA CTA GGT GAT ATC GAG C/ ACC AGA CAA GAG TTT AAG AGA TAT GTA TC |
| Reprogramming virus: | KOS | ATG CAC CGC TAC GAC GTG AGC GC/ ACC TTG ACA ATC CTG ATG TGG |
| Reprogramming virus: | Klf4 | TTC CTG CAT GCC AGA GGA GCC C/ AAT GTA TCG AAG GTG CTC AA |
| Reprogramming virus: | c-Myc | TAA CTG ACT AGC AGG CTT GTC G/ TCC ACA TAC AGT CCT GGA TGA TGA TG |
Reprogramming the fibroblasts
Fibroblasts were thawed and cultivated in human fibroblast medium. When ~80% confluent, the MZT04 cells were transfected with Sendai virus (SeV) based non-integration CytoTune™ iPS Reprogramming Kit (Life Technologies) (4) according to manufacturer’s instructions. Colonies began to appear after 11 days and were picked after three weeks. Three colonies were manually picked and expanded onto mouse embryonic fibroblast feeder cells in hiPSC media (DMEM/F-12 (Life Technologies), 20% KSR (Life Technologies), 10 ng/mL bFGF (R & D Systems), 1% nonessential amino acids (Life Technologies), 1% Penicillin-Strepromyocin-Glutamine (Gibco), Primocin™ (Invivogen), and 0.1 mM β-mercaptoethanol (Sigma))
Flow cytometry
Single cell suspension was obtained using 0.05% Tryspin (Gibco). hiPSCs were then resuspended in PBS with 1% BSA. Antibody incubation lasted 30 min at 4 °C with conjugated antibodies (Tables 2 and 3).
Karyotyping and STR analysis
The four lines were karyotyped using metaphase spreads and G-banding by Cell Line Genetics (Madison, WI). Cell Line Genetics also performed Identity analysis on the four cell lines using the PowerPlex 16 System (cat# DC6531, Promega) (Table 2).
Teratoma formation assay
All animal work was first approved by the UCLA Office of Animal Research Oversight. Cells were digested with 1 mg/mL collagenase IV (Life Technologies), and re-suspended in cold Matrigel (Corning). Cells were then injected into the left and right testis of n = 2 CB17/-Prkdcscid Lystbg/Cr (scid beige) mice using survival surgery. Each testis was injected with 40 μL of matrigel containing ~ 1.5 × 106 hiPSCs cells in small clumps. Two months after the transplant, the testes containing tumors were removed and fixed using 4% Paraformaldehyde (PFA). The excised tumors were stained for hematoxylin and eosin (H&E) (Table 2).
PluriTest assay
Cryopreserved pelleted cells were sent to Thermo Fisher Scientific. Transcriptional profiles of the hiPSC lines were compared to an extensive reference set of previously characterized pluripotent stem cell lines. The Pluripotency score is an indication of how strongly a model-based pluripotency signature is expressed in the pelleted cells. The novelty score indicates the general model fit for a given sample.
Immunofluorescence staining
Immunofluorescence staining was performed by fixing the hiPSCs in 4%PFS for 15 min at room temperature, and then permeabilizing the cells with PBS plus 0.5% Triton™ X-100 (Sigma). The hiPSCs were then blocked in 10% donkey serum (Jackson Immunoresearch) for 30 min at room temperature. Cells were incubated overnight at 4 °C with primary antibodies and then were incubated in secondary antibodies for 1 h. Immunofluorescence was imaged using a Zeiss LSM 880 confocal laser-scanning microscope (Table 2 and 3).
Absence of the reprogramming virus
RNA was isolated according to manufacturer’s instructions (cytotune) (FUSAKI et al., 2009) from reprogrammed fibroblasts at P0 before hiPSCs were picked and cultured. cDNA was synthesized from the RNA and RT-PCR was performed using primers provided from the manufacturer (FUSAKI et al., 2009) (Table 3).
Mycoplasma detection
Mycoplasma was regularly tested using MycoAlert kit from Lonza Catalog #LT07–318.
Key resources table
| Unique stem cell lines identifier | UCLAi001-A |
|---|---|
| UCLAi001-B | |
| UCLAi001-C | |
| Alternative names of stem cell lines | MZT04D |
| MZT04J | |
| MZT04C | |
| Institution | UCLA |
| Contact information of distributor | Dr. Amander Clark |
| Type of cell lines | hiPSC |
| Origin | Human |
| Cell Source | Fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Sendai |
| Multiline rationale | Isogenic clones |
| Gene modification | No |
| Type of modification | N/A |
| Associated disease | None |
| Gene/locus | N/A |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | N/A |
| Cell line repository/bank | N/A |
| Ethical approval | UCLA Office of the Human Resource Protection Program- IRB#16-001176-CR-00002 and UCLA Embryonic Stem Cell Research Oversight Committee (ESCRO# 20016-003) |
Acknowledgements
We are appreciative of the MCDB/BSCRC Imaging Core, BSCRC Flow cytometry core and BSCRC Genomics core. We also thank Jessica Scholes, Felicia Codrea and Jeffrey Calimlim of the UCLA BSCRC FACS core. In addition, we are grateful to Tsotne Chitiashvili for conducting the mycoplasma testing, and Rachel Kim for teratoma assays. Dr. Erica Pandolfi is a postdoctoral fellow supported by UPLIFT: UCLA Postdocs’ Longitudinal Investment in Faculty (Award # K12 GM106996). This project was funded by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Innovation Award (ATC). We also gratefully acknowledge funds from an anonymous donor to support this work.
References
Aalto-Setälä et al., 2009K. Aalto-Setälä, B.R. Conklin, B. Lo Obtaining consent for future research with induced pluripotent cells: opportunities and challenges PLoS Biol., 7 (2009), Article e1000042, 10.1371/journal.pbio.1000042
FUSAKI et al., 2009N. Fusaki, H. Ban, A. Nishiyama, K. Saeki, M. Hasegawa Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome Proc. Japan Acad. Ser. B, 85 (2009), pp. 348-362, 10.2183/pjab.85.348
Müller et al., 2011F.J. Müller, B.M. Schuldt, R. Williams, D. Mason, G. Altun, E.P. Papapetrou, S. Danner, J.E. Goldmann, A. Herbst, N.O. Schmidt, J.B. Aldenhoff, L.C. Laurent, J.F. Loring A bioinformatic assay for pluripotency in human cells Nat. Methods, 8 (2011), pp. 315-317, 10.1038/nmeth.1580
Tokusumi et al., 2002T. Tokusumi, A. Iida, T. Hirata, A. Kato, Y. Nagai, M. Hasegawa Recombinant sendai viruses expressing different levels of a foreign reporter gene Virus Res., 86 (2002), pp. 33-38, 10.1016/S0168-1702(02)00047-3


